Research
Role of Inhibitor of DNA binding genes in the Regulation of the Mammalian Circadian Clock
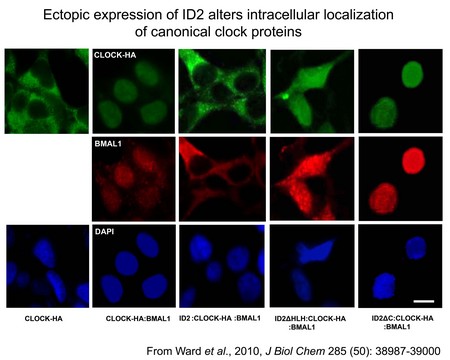
The molecular circadian clock consists of an autoregulatory transcriptional-translational feedback loop composed of positive and negative regulators. Work over the last 14 years has identified at least nine such components, but additional genes and modifiers are being identified. In addition, most tissues of the body harbor cell-autonomous circadian clocks. One such additional gene, identified in my laboratory using a DNA microarray screen, is the transcriptional inhibitor Inhibitor of DNA-binding 2 (ID2). It is rhythmically expressed in the master clock structure in the hypothalamic brain known as the suprachiasmatic nucleus (SCN), and throughout the body in various peripheral tissues (e.g. heart and liver). Current studies are to evaluate the role of genes such as Id2 in the organization of the central oscillator, and to identify novel molecules relevant to cellular clock function. We are also interested in understanding how light resets the molecular clock (input), and how the clock regulates down-stream clock-controlled genes (i.e. output, hands of the clock). The lab is using continuous activity monitoring to identify behavioral phenotypes in transgenic mouse models (e.g. Id2 knockout mice) that are maintained under a variety of photocycle conditions, and exposed to artificial time-zone changes and acute light/pharmacologic/behavioral treatments. Results thus far have revealed that in the absence of the Id2 gene, mice adapt to large time-zone changes (e.g. mimicking a flight from Berlin to Los Angeles) more rapidly than wildtype individuals. We are also using real-time monitoring of clock gene expression in tissues and cells derived from transgenic mice that express Firefly luciferase in a rhythmic manner. We are using DNA microarray and real-time quantitative RT-PCR analyses to identify and characterize clock regulated genes in brain and peripheral tissues (e.g. liver and heart); tissue culture of immortalized fibroblasts that exhibit circadian oscillations in gene expression as a model of the in vivo rodent circadian clock; protein interaction analyses (e.g. western blot, co-immunoprecipitation, 2-hybrid, immunofluorescence) to understand interaction dynamics between clock related proteins; and traditional neuroanatomical techniques (e.g. in situ hybridization, immunohistochemistry, neuronal track tracing) to characterize clock gene function in the brain. Furthermore, we have begun to utilize Drosophila fruit fly as a tractable system in which to examine the role of Id genes further within the circadian system.
Role of Id2 in metabolic regulation
We are interested in the relationship between the circadian clock and lipid and glucose metabolism. We have demonstrated that Id2 null mice exhibit a metabolic phenotype characterized by low lipid storage (lean body form, low quantity of white adipose tissue, and reduced lipid levels in liver) and disturbances in rhythms in genes expressed in the liver that are associated with daily changes in lipid and glucose availability and utilization. Current studies are to evaluate the role of genes such as Id2 in the pathways through which the circadian clock regulates 24 hr rhythms in metabolic function.
Circadian Genomics of Anopheles gambiae
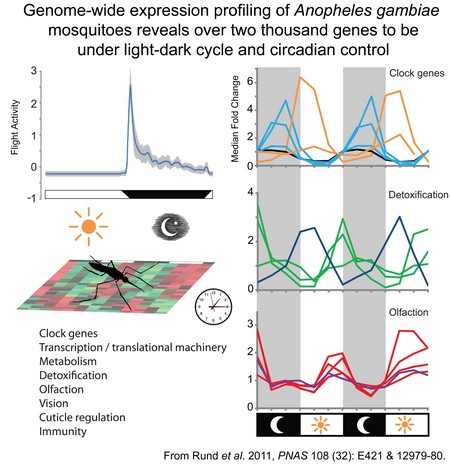
The molecular basis for circadian rhythms has been investigated in model organisms such as the mouse and Drosophila, but little is known of circadian regulation in disease vector species, such as the Anopheles mosquito (malaria vector). The mosquito exhibits a wealth of circadian behavior, such as feeding and breeding activity, but almost nothing is know of how the circadian system in this insect family are comprised and how the core molecular clock components regulate physiology and behavior. This research examines the circadian clock of Anopheles gambiae, a vector of the malaria parasite in Africa. We are interested in 1) understanding how the circadian clock is comprised at a molecular and cellular level; and 2) defining the circadian control of the transcriptome and to elucidate pathways that interconnect with overt rhythmic aspects of the species behavior and physiology. Research in the laboratory emphasizes techniques of molecular biology, genetics, and genomics, including high density DNA microarray analysis. We have discovered that genes associated with processes of metabolism, detoxification and sensory modalitities of vision and olfaction are under rhythmic control. Characterization of the circadian regulation of the mosquito transcriptome will allow us to better understand both insecticide resistance in this species, as well as generate improved understanding of temporal gated behavior such as time of blood feeding. Furthermore, examination of A. gambiae sub-species that exhibit differences in their circadian behavior, physiological responses and resistance to insecticides, will allow us to identify specific underlying molecular pathways that correlate with the variants observed in the overt rhythms.
Anopheles gambiae and Aedes aegypti mosquito gene expression data
